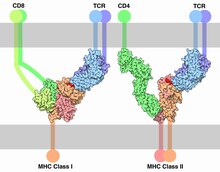
T-cell receptor
[5] In 1982, Nobel laureate James P. Allison first discovered a clonally expressed T-cell surface epitope in murine T lymphoma.[7][8] Then, Tak Wah Mak[9] and Mark M. Davis[10] identified the cDNA clones encoding the human and mouse TCR respectively in 1984.These findings allowed the entity and structure of the elusive TCR, known before as the "Holy Grail of Immunology", to be revealed.HV4 of the β-chain is not thought to participate in antigen recognition as in classical CDRs, but has been shown to interact with superantigens.Unlike immunoglobulins, however, TCR genes do not undergo somatic hypermutation, and T cells do not express activation-induced cytidine deaminase (AID).It is the unique combination of the segments at this region, along with palindromic and random nucleotide additions (respectively termed "P-" and "N-"), which accounts for the even greater diversity of T-cell receptor specificity for processed antigenic peptides.In the plasma membrane the TCR receptor chains α and β associate with six additional adaptor proteins to form an octameric complex.Charged residues in the transmembrane domain of each subunit form polar interactions allowing a correct and stable assembly of the complex.[20][21] To do so, T cells have a very high degree of antigen specificity, despite the fact that the affinity to the peptide/MHC ligand is rather low in comparison to other receptor types.[25] A negative correlation between the dissociation rate of the pMHC-TCR complex and the strength of the T-cell response has been observed.[26] That means, pMHC that bind the TCR for a longer time initiate a stronger activation of the T cell.The occupational model simply suggests that the TCR response is proportional to the number of pMHC bound to the receptor.Given this model, a shorter lifetime of a peptide can be compensated by higher concentration such that the maximum response of the T cell stays the same.[32] The essential function of the TCR complex is to identify specific bound antigen derived from a potentially harmful pathogen and elicit a distinct and critical response.The signal transduction mechanism by which a T cell elicits this response upon contact with its unique antigen is termed T-cell activation.[33] On a population level, T-cell activation depends on the strength of TCR stimulation, the dose–response curve of ligand to cytokine production is sigmoidal.The robust sigmoid dose-response curve on population level results from individual T cells having slightly different thresholds.These co-stimulatory receptors are expressed only when an infection or inflammatory stimulus is detected by the innate immune system, Known as a "Danger signal".This two-signal system makes sure that T cells only respond to harmful stimuli (i.e. pathogens or injury) and not to self-antigens.[33] There are a myriad of molecules involved in the complex biochemical process (called trans-membrane signaling) by which T-cell activation occurs.Once the TCR binds a specific pMHC, the tyrosine residues of the immunoreceptor tyrosine-based activation motifs (ITAMs) in its CD3 adaptor proteins are phosphorylated.[39] Molecules that bind the LAT/Slp76 complex include: Phospholipase Cγ1 (PLCγ1), SOS via a Grb2 adaptor, Itk, Vav, Nck1 and Fyb.It hydrolyses PIP2 into two secondary messenger molecules, namely the membrane-bound diacyl glycerol (DAG) and the soluble inositol 1,4,5-trisphosphate (IP3).Transcription factors involved in T-cell signaling pathway are the NFAT, NF-κB and AP1, a heterodimer of proteins Fos and Jun.[33] Additionally, there is evidence that PI-3K via signal molecules recruits the protein kinase AKT to the cell membrane.CARMA1 then undergoes a conformational change which allows it to oligomerize and bind the adapter proteins BCL10, CARD domain and MALT1.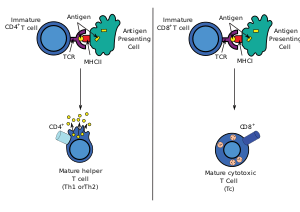
OPM superfamilyMembranomeAntigen presentation"cytotoxic" CD8+ cells"helper" CD4+ cellsChr. 14Chr. 7protein complexT cellsantigenmajor histocompatibility complexdegenerateheterogamma and deltaontogenyleukemiaOrthologuespolypeptidessignal transductiontranscription factorsnon-catalytic tyrosine-phosphorylated receptorsJames P. AllisonPhilippa MarrackJohn KapplerTak Wah MakMark M. Daviscancer immunotherapycheckpoint inhibitionγδ T cellsImmunoglobulin superfamilydomainβ-sheetscomplementarity-determining regionsframework regionprocessed antigenN-terminalC-terminalsuperantigensantibodiesB-cell antigen receptorsgenetic recombinationsomatic V(D)J recombinationimmunoglobulinssomatic hypermutationactivation-induced cytidine deaminaselymphocytesthymusbone marrowantigen-binding siteVJ recombinationpalindromicCDR loopsTCR revisioncytoplasmicnon-catalytic tyrosine-phosphorylated receptorimmunoreceptor tyrosine-based activation motifMHC IIMHC class IIantigen-presenting cellsMHC class Idissociation constantsurface plasmon resonancekinetic proofreadingantigen presenting celldose–response curveco-stimulatory receptorscytokinestrans-membrane signalingNTR receptor familyimmunoreceptor tyrosine-based activation motifsSrc kinaseco-receptorhelper T cellsregulatory T cellscytotoxic T cellsphosphatasekinetic segregationSH2 domainscaffold proteinSlp-76Phospholipase CPLCγ1second messengerphosphatidylinositol (3,4,5)-trisphosphatephosphoinositide 3-kinasephosphatidylinositol 4,5-bisphosphatesecondary messengerdiacyl glycerolinositol 1,4,5-trisphosphateNF-κBheterodimerinterleukin-2calcium signalingcalcium channel receptorsendoplasmic reticulumcalmodulincalcineurinnucleusnuclear localization sequenceprotein kinase CCARMA1CARD domainubiquitin ligaseUbiquitinationIκB kinaseAP1 factorMAPK signaling pathwaysGTPaseRasGRPguanine nucleotide exchange factorguanosine diphosphateguanosine triphosphateB-cell receptorCo-stimulationImmTACMHC multimerBibcodeMedical Subject HeadingsTransmembrane receptorsimmune receptorsFc receptorFcεRIFcεRIIC-type lectinFcγRIFcγRII