Model lipid bilayer
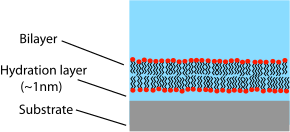
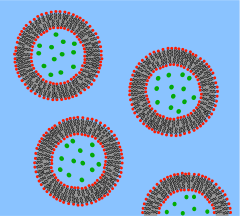
Supported bilayers are anchored to a solid substrate, increasing stability and allowing the use of characterization tools not possible in bulk solution.This annulus is required to maintain stability by acting as a bridge between the ~5 nm bilayer and the tens of micrometers thick sheet in which the aperture is made.[4] Black lipid membranes are also well suited to electrical characterization because the two chambers separated by the bilayer are both accessible, allowing simple placement of large electrodes.[6][7] This modification of the patch clamp technique enables low noise recording, even at high potentials (up to 600 mV), at the expense of additional preparation time.SLBs will remain largely intact even when subject to high flow rates or vibration and, unlike black lipid membranes, the presence of holes will not destroy the entire bilayer.[11] Another advantage of the supported bilayer is that, because it is on a flat hard surface, it is amenable to a number of characterization tools which would be impossible or would offer lower resolution if performed on a freely floating sample.More recently, AFM has also been used to directly probe the mechanical properties of single bilayers[15] and to perform force spectroscopy on individual membrane proteins.[16] These studies would be difficult or impossible without the use of supported bilayers since the surface of a cell or vesicle is relatively soft and would drift and fluctuate over time.[17] Dual polarisation interferometry is a high resolution optical tool for characterising the order and disruption in lipid bilayers during interactions or phase transitions providing complementary data to QCM measurements.But unlike AFM, NSOM uses an optical rather than physical interaction with the sample, potentially perturbing delicate structures to a lesser extent.[23][24] Later work extended this concept by integrating microfluidics to demonstrate that stable composition gradients could be formed in bilayers,[25] potentially allowing massively parallel studies of phase segregation, molecular binding and cellular response to artificial lipid membranes.Unwanted substrate interactions are a much greater problem when incorporating integral membrane proteins, particularly those with large domains sticking out beyond the core of the bilayer.In these systems the bilayer is supported on a loose network of hydrated polymers or hydrogel which acts as a spacer and theoretically prevents denaturing substrate interactions.Additionally the spacer layer creates an ionic reservoir[36] that readily enables ac electrical impedance measurement across the bilayer.[42] Dual polarisation interferometry can measure unilamelar and multilamelar structures and insertion into and disruption of the vesicles in a label free assay format.[45] Compared to supported bilayers, GUVs present a more “natural” environment since there is no rigid surface that might induce defects, affect the properties of the membrane or denature proteins.A variety of methods exist to encapsulate proteins or other biological reactants within such vesicles, making GUVs an ideal system for the in vitro recreation (and investigation) of cell functions in cell-like model membrane environments.In aqueous solutions, micelles are assemblies of amphipathic molecules with their hydrophilic heads exposed to solvent and their hydrophobic tails in the center.Nanodiscs are more stable than bicelles and micelles at low concentrations, and are very well-defined in size (depending on the type of protein coat, between 10 and 20 nm).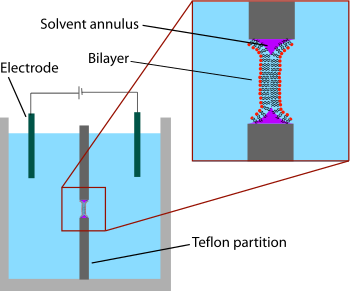


Membrane modelsbilayerin vitrocell membranesnucleussynthetic biologyartificial cellslipidscharacterization toolsmembrane proteinsTeflonhydrophobic solventpartition coefficientdecanesqualeneinterferesvoltage gated ion channelsion channelsdetergentpatch clampchloroformlong-term experimentsAtomic force microscopyphase separationmechanical propertiesquartz crystal microbalanceDual polarisation interferometryphase transitionsEvanescent fieldtotal internal reflection fluorescence microscopysurface plasmon resonanceinterferometric scattering microscopynear field scanning optical microscopymicrofluidicssynapticzwitterionicsilicadenaturedhydrogeldisulphideself assembled monolayerselectrical impedanceUnilamellar liposomex-ray diffractionfusionlipid raftsmicellesamphipathicNanodiscsBibcodeDyson HJSelf-replicatingCellular lifeBacteriaArchaeaEukaryotaAnimaliaPlantaeProtistaIncertae sedisParakaryonBiological dark matterGiant virusdsRNA virusssRNA-RT virusdsDNA-RT virusSubviralagentsViroidPospiviroidaeAvsunviroidaeHelper-virusdependentSatelliteVirophageVirusoidDefective interfering particleMammalian prionFungal prionNucleic acidMobile geneticelementsMobilomeHorizontal gene transferGenomic islandTransposable elementClass I or retrotransposonClass II or DNA transposonPlasmidFertilityResistanceVirulenceCosmidFosmidPhagemidGroup I intronGroup II intronRetrozymeDNA replicationRNA replicationChromosomeLinearCircularExtrachromosomal DNASecondary chromosomeGenomeGene duplicationNon-coding DNAOrigin of replicationRepliconEndogenous viral elementProvirusProphageEndogenous retrovirusTranspovironRepeated sequences in DNATandem repeatInterspersed repeatEndosymbiosisMitochondrionMitosomeHydrogenosomePlastidChloroplastChromoplastGerontoplastLeucoplastApicoplastKappa organism