ChIP sequencing
ChIP-seq combines chromatin immunoprecipitation (ChIP) with massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins.Determining how proteins interact with DNA to regulate gene expression is essential for fully understanding many biological processes and disease states.[3] As an alternative to the dependence on specific antibodies, different methods have been developed to find the superset of all nucleosome-depleted or nucleosome-disrupted active regulatory regions in the genome, like DNase-Seq[4] and FAIRE-Seq.The ChIP process enhances specific crosslinked DNA-protein complexes using an antibody against the protein of interest followed by incubation and centrifugation to obtain the immunoprecipitation.Oligonucleotide adaptors are then added to the small stretches of DNA that were bound to the protein of interest to enable massively parallel sequencing.[20] Inferring regulatory network: ChIP-seq signal of Histone modification were shown to be more correlated with transcription factor motifs at promoters in comparison to RNA level.[21] Hence author proposed that using histone modification ChIP-seq would provide more reliable inference of gene-regulatory networks in comparison to other methods based on expression.STAT1 experimental ChIP-seq data have a high degree of similarity to results obtained by ChIP-chip for the same type of experiment, with greater than 64% of peaks in shared genomic regions.ChIP-seq also has the potential to detect mutations in binding-site sequences, which may directly support any observed changes in protein binding and gene regulation.One of the most popular methods[citation needed] is MACS which empirically models the shift size of ChIP-Seq tags, and uses it to improve the spatial resolution of predicted binding sites.A mathematical more rigorous method BCP (Bayesian Change Point) can be used for both sharp and broad peaks with faster computational speed,[23] see benchmark comparison of ChIP-seq peak-calling tools by Thomas et al.[24] Another relevant computational problem is differential peak calling, which identifies significant differences in two ChIP-seq signals from distinct biological conditions.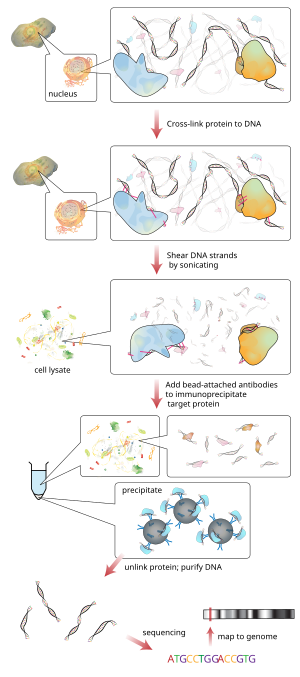
proteinchromatin immunoprecipitationmassively parallelDNA sequencingbinding sitesChIP-on-chiptranscription factorsphenotypegene expressionepigeneticgenotypeChIP-chiphybridization arraychromatinpolymerasestranscriptional machineryprotein modificationsnucleosomeDNase-SeqFAIRE-Seqcross-linkingantibodyOligonucleotidemassively parallel sequencingtiling arrayssequencing-by-synthesismicroarraytranscription enhancersHistone modificationpeak callingHidden Markov ModelsChIP-PETMammalian promoter databaseCUT&RUN sequencingCUT&Tag sequencingSono-SeqHITS-CLIPCLIP-SeqPAR-CLIPRIP-ChipCompetition-ChIPChiRP-SeqChIP-exoDRIP-seqTCP-seqBibcodeChIP-SeqWayback Machine