Agrobacterium tumefaciens
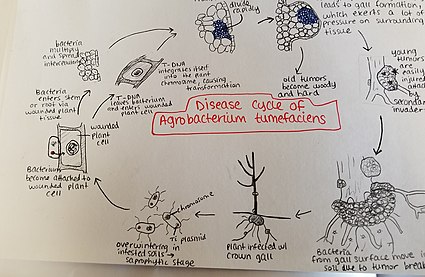
[2] Agrobacterium tumefaciens[3][2] (also known as Rhizobium radiobacter) is the causal agent of crown gall disease (the formation of tumours) in over 140 species of eudicots.[10] Economically, A. tumefaciens is a serious pathogen of walnuts, grape vines, stone fruits, nut trees, sugar beets, horse radish, and rhubarb, and the persistent nature of the tumors or galls caused by the disease make it particularly harmful for perennial crops.In the presence of opines, A. tumefaciens produces a diffusible conjugation signal called N-(3-oxo-octanoyl)-L-homoserine lactone (3OC8HSL) or the Agrobacterium autoinducer.Following an initial weak and reversible attachment, the bacteria synthesize cellulose fibrils that anchor them to the wounded plant cell to which they were attracted.[citation needed] After production of cellulose fibrils, a calcium-dependent outer membrane protein called rhicadhesin is produced, which also aids in sticking the bacteria to the cell wall.The effect of different parameters such as infection time, acetosyringone, DTT, and cysteine have been studied in soybean (Glycine max).When acetosyringone and other substances are detected, a signal transduction event activates the expression of 11 genes within the VirB operon which are responsible for the formation of the T-pilus.The structure of the T-pilus showed that the central channel of the pilus is too narrow to allow the transfer of the folded VirD2, suggesting that VirD2 must be partially unfolded during the conjugation process.To cause gall formation, the T-DNA encodes genes for the production of auxin or indole-3-acetic acid via the IAM pathway.The T-DNA contains genes for encoding enzymes that cause the plant to create specialized amino acid derivatives which the bacteria can metabolize, called opines.The Asilomar Conference in 1975 established widespread agreement that recombinant techniques were insufficiently understood and needed to be tightly controlled.[27][28] The DNA transmission capabilities of Agrobacterium have been vastly explored in biotechnology as a means of inserting foreign genes into plants.[citation needed] Natural genetic transformation in bacteria is a sexual process involving the transfer of DNA from one cell to another through the intervening medium, and the integration of the donor sequence into the recipient genome by homologous recombination.Once the bacteria have entered the plant, they occur intercellularly and stimulate surrounding tissue to proliferate due to cell transformation.This secondary invasion causes the breakdown of the peripheral cell layers as well as tumor discoloration due to decay.Breakdown of the soft tissue leads to release of the Agrobacterium tumefaciens into the soil allowing it to restart the disease process with a new host plant.[35] It is recommended that infected plant material be burned rather than placed in a compost pile due to the bacteria's ability to live in the soil for many years.This method, which was successful at controlling the disease on a commercial scale, had the risk of K84 transferring its resistance gene to the pathogenic Agrobacteria.Agrobacteria have the widest host range of any plant pathogen,[41] so the main factor to take into consideration in the case of crown gall is environment.Consequently, in exceptionally harsh winters, it is common to have an increased incidence of crown gall due to the weather-related damage.
- Agrobacterium cell
- Agrobacterium chromosome
- Ti Plasmid (a. T-DNA, b. vir genes, c. replication origin, d. opines catabolism)
- Plant cell
- Plant mitochondria
- Plant chloroplast
- Plant nucleus
- VirA recognition
- VirA phosphorylates VirG
- VirG causes transcription of Vir genes
- Vir genes cut out T-DNA and form nucleoprotein complex ("T-complex")
- T-complex enters plant cytoplasm through T-pilus
- T-DNA enters into plant nucleus through nuclear pore
- T-DNA achieves integration


Scientific classificationBacteriaPseudomonadotaAlphaproteobacteriaHyphomicrobialesRhizobiaceaeAgrobacteriumBinomial nameType strainSynonymsAgrobacterium radiobactertumourseudicotsGram-negativebacteriumgenomeT-DNA binary vectorsAlphaproteobacteriumnitrogen-fixinglegumesymbiontspathogenicwalnutsgrape vinesstone fruitssugar beetshorse radishrhubarbvirulenttumour-inducing plasmidrhizospherebacterial conjugationopinestranscription factortranscriptionflagellaphotoassimilatesstrainschemotacticallyacetosyringoneVirA proteinkinasephosphorylatesoperonscellulosefibrilsmicrocolonycalcium-dependentproteinHomologuesAgrobacterium-mediated transformationCatecholFerulic acidGallic acidp-Hydroxybenzoic acidProtocatechuic acidPyrogallic acidResorcylic acidSinapinic acidSyringic acidVanillinplant cellsignal transductionoperonpolypeptidepeptide bondplasma membraneYeast two-hybridencodeATPaseactive transportNuclear localization signalsnuclear pore complexnucleuschromatincytokininsenzymesamino acidmetabolizenopalineTi plasmidspathovarplasmidschromosomevirulenceAgroinfiltrationplant tissue culturesAsilomar ConferencebiotechnologyMarc Van MontaguJeff Schellgenetic engineeringluciferaseluminescencereporter geneArabidopsis thalianatransgenicpathogensproteinsquorum sensingNatural genetic transformationhomologous recombinationRhizobium rhizogenesdeletion mutantInt J Syst Evol MicrobiolScienceBibcodeInternational Bulletin of Bacteriological Nomenclature and TaxonomyInt J Syst BacteriolApplied and Environmental MicrobiologyMissouri Botanical GardenCurrent Protocols in MicrobiologyMicrobiology SpectrumMicrobiology and Molecular Biology ReviewsThe Plant JournalCiteSeerXWashington, D.C.National Academies PressBioScienceProceedings of the National Academy of Sciences of the United States of AmericaHilgardiaAmsterdamElsevier Academic PressUniversity of Minnesota ExtensionFunctional Plant BiologyMolecular Plant Pathology